The CRL has developed a mathematical framework for the generation of an unbiased, deformable, spatiotemporal atlas of the fetal brain from magnetic resonance imaging (MRI) of normal fetuses scanned prenatally. Our atlas serves to capture the inter-subject anatomic variability of the fetal brain over the fetal brain growth period (Gholipour et al., 2017) and is currently available between 21 weeks gestational age to 38 weeks. The atlas has been constructed following an unbiased minimum distance template estimation approach which utilizes symmetric diffeomorphic deformation and the cross-correlation similarity metric integrated with kernel regression in age.
This methodology is characterized by its sharpness, which indicates superior inter-subject registration performance as compared to other schemes, such as those that have employed affine registration, Demons registration, or free-form deformation. In result, this atlas can be effectively used in spatial normalization (inter-subject image registration), fetal brain localization and matching, and atlas-based segmentation.
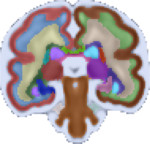
 |
|---|
| Fetal Brain Atlas weeks 21 through 37 |
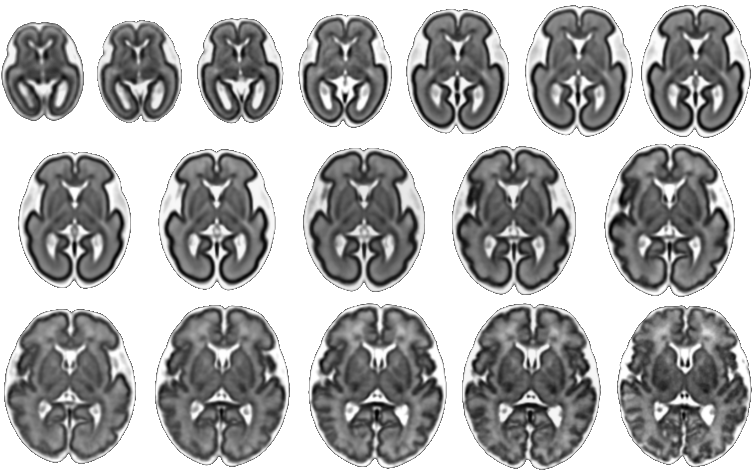
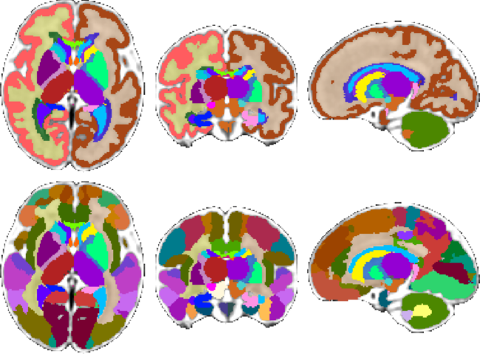 |
|---|
| Fetal Brain Atlas tissue and regional segmentations |
For these purposes, the atlas comes with brain tissue and structure labels including cortical gray matter, white matter, subcortical gray matter structures, CSF, lateral ventricles, brainstem, and cerebellum labels, and more. In addition, gestational age weeks 21-30 include labels for developing white matter layers, including the subplate, intermediate zone, and ventricular zone. An update of the atlas, including a week 38 template and an additional regionally divided cortex parcellation was released in February 2018. Future plans include expanding the atlas to include earlier gestational ages. Please check back for updates.
Potential applications of the fetal brain atlas are the development of faster, more accurate, automated methods of MRI structural analysis, and disease biomarker identification in utero. Assessing the volume and overall morphology of the fetal ventricles, identifying and segmenting the transitory fetal subplate zone, and developing a method of automatic fetal MRI brain localization are some of our ongoing research projects.
Fetal Diffusion Atlas
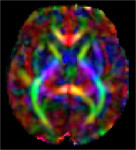 |
|---|
| Fetal Diffusion Atlas Color-FA map, 35 weeks gestational age |
We have released a Fetal Diffusion Tensor Imaging (DTI) Atlas, available through the download link below. This atlas, the first of its kind to be created from in-utero fetal images, is comprised of composite tensor maps generated from 67 DTI scans of developing fetuses in normative pregnancies. The resulting fiber connections characterized by the atlas were validated through comparison with existing data of ex-vivo and in-utero fetuses, as well as pre-term infants. The atlas release includes fractional anisotropy (FA), color fractional anisotropy (CFA), and Mean Diffusivity (MD) maps for gestational age weeks 22 through 38.
Link to download Fetal Brain Atlas
References
- A Gholipour, CK Rollins, C Velasco-Annis, A Ouaalam, A Akhondi-Asl, O Afacan, C Ortinau, S Clancy, C Limperopoulos, E Yang, JA Estroff, and SK Warfield. A normative spatiotemporal MRI atlas of the fetal brain for automatic segmentation and analysis of early brain growth, Scientific Reports 7, Article number: 476 (2017). http://www.nature.com/articles/s41598-017-00525-w
- A Gholipour, C Limperopoulos, S Clancy, C Clouchoux, A Akhondi-Asl, J A Estroff, and S K Warfield. Construction of a Deformable Spatiotemporal MRI Atlas of the Fetal Brain: Evaluation of Similarity Metrics and Deformation Models. MICCAI 2014. <LINK TO Gholipour MICCAI 2014 paper1126.pdf>
- S Khan, L Vasung, B Marami, CK Rollins, O Afacan, C Ortinau, E Yang, SK Warfield, and A Gholipour. Fetal Brain Growth Portrayed by a Spatiotemporal Diffusion Tensor MRI Atlas Computed From In Utero Images. NeuroImage 2018. https://doi.org/10.1016/j.neuroimage.2018.08.030
Other References
- B Marami, SSM Salehi, O Afacan, B Scherrer, CK Rollins, E Yang, J Estroff, SK Warfield, and A Gholipour. Temporal slice registration and robust diffusion-tensor reconstruction for improved fetal brain structural connectivity analysis. NeuroImage (2017). https://www.sciencedirect.com/science/article/pii/S1053811917303282
- SSM Salehi, D Erdogmus, and A Gholipour. Auto-context convolutional neural network (auto-net) for brain extraction in magnetic resonance imaging. IEEE Transactions on Medical Imaging (2017). https://arxiv.org/pdf/1703.02083.pdf
- SSM Salehi, SR Hashemi, C Velasco-Annis, A Ouaalam, J Estroff, D Erdogmus, SK Warfield, and A Gholipour. Real-Time Automatic Fetal Brain Extraction in Fetal MRI by Deep Learning. IEEE International Symposium on Biomedical Imaging (ISBI 2018). https://arxiv.org/abs/1710.09338
- SSM Salehi, S Khan, D Erdogmus, and A Gholipour. Real-time Deep Pose Estimation with Geodesic Loss for Image-to-Template Rigid Registration. IEEE Transactions on Medical Imaging (2018). https://ieeexplore.ieee.org/abstract/document/8443391
- S Khan, CK Rollins, C Ortinau, O Afacan, SK Warfield, and A Gholipour. Tract-Specific Group Analysis in Fetal Cohorts using in utero Diffusion Tensor Imaging. MICCAI 2018. https://link.springer.com/chapter/10.1007/978-3-030-00931-1_4